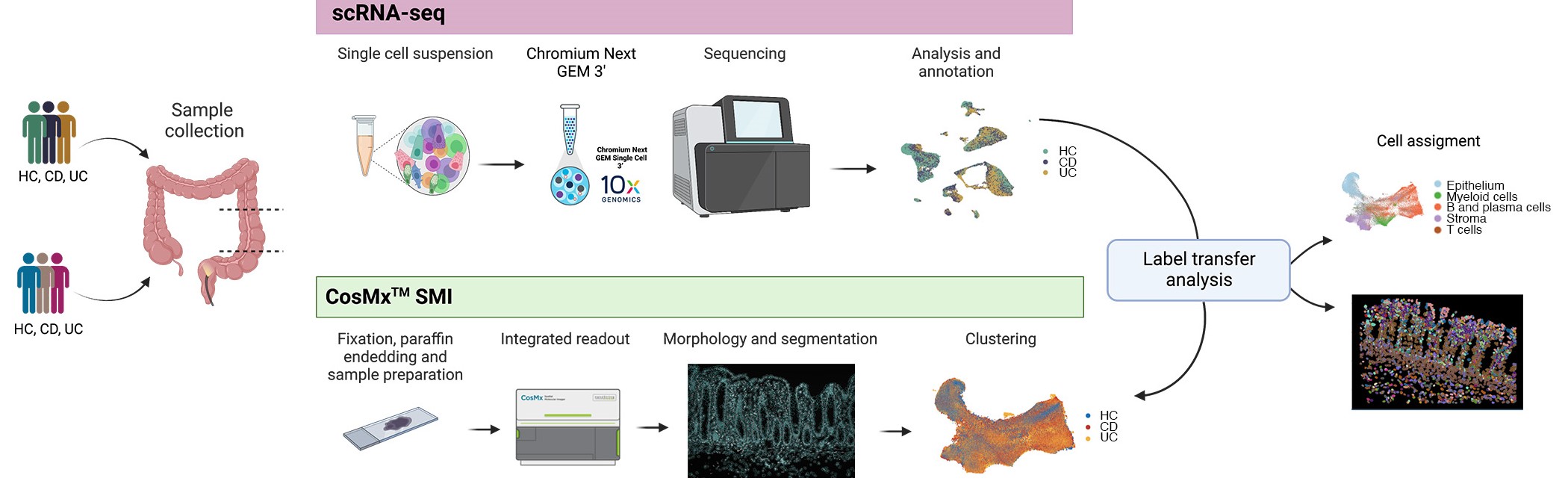
Ulcerative colitis (UC) and Crohn’s disease (CD) are chronic inflammatory intestinal diseases that show a perplexing heterogeneity in manifestations and response to treatment. The molecular basis for this heterogeneity remains uncharacterized. We applied single-cell RNA sequencing and CosMx™ Spatial Molecular Imaging to human colon and found the highest diversity in cellular composition in the myeloid compartment of UC and CD patients.
Besides resident macrophage subsets (M0 and M2), patients showed a variety of activated macrophages including classical (M1 CXCL5 and M1 ACOD1) and new inflammation-dependent alternative (IDA) macrophages. In addition, we captured intestinal neutrophils in three transcriptional states. Subepithelial IDA macrophages expressed NRG1, which promotes epithelial differentiation. In contrast, NRG1low IDA macrophages were expanded within the submucosa and in granulomas, in proximity to abundant inflammatory fibroblasts, which we suggest may promote macrophage activation.
We conclude that macrophages sense and respond to unique tissue microenvironments, potentially contributing to patient-to-patient heterogeneity.